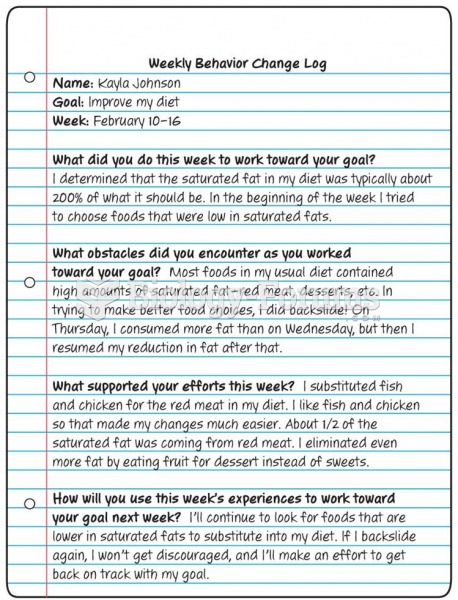
Definition for Loss of heterozygosity
From Biology Forums Dictionary
Most diploid cells, for example human somatic cells, contain two copies of the genome, one from each parent (chromosome pair); each copy contains approximately 3 billion bases (Adenine (A), Guanine (G), Cytosine (C) or Thymine (T)). For the majority of positions in the genome the base present is consistent between individuals, however a small percentage may contain different bases (usually one of two for instance ‘A’ or ‘G’) and these positions are called ‘single nucleotide polymorphisms’ or ‘SNPs’. When the genomic copies derived from each parent have different bases for these polymorphic regions (SNPs) the region is said to be heterozygous. The majority of the genome within somatic cells of individuals are heterozygous. However, one parental copy of a region can sometimes be lost, resulting in the region lacking differences at these polymorphic loci (SNPs) and therefore showing loss of heterozygosity (LOH). Loss of heterozygosity due to loss of one parental copy in a region is also called hemizygosity in that region.